Computational Studies of Dipyridodiazepinones as Human Immunodeficiency Virus ( HIV) Reverse Transcriptase Inhibitors
Reiner G. Villavicencio and Junie B. Billones*
Department of Physical Sciences and Mathematics, College of Arts and Sciences,
University of the Philippines Manila, Padre Faura St., Ermita, Manila 1000 Philippines
*Corresponding author: This email address is being protected from spambots. You need JavaScript enabled to view it.
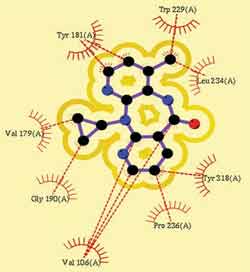
Hydrophobic interactions of nevirapine with aromatic and aliphatic amino acid residues of the wild type reverse transcriptase.
ABSTRACT
A series of dipyridodiazepinone derivatives that are highly active against wild-type and K103N – Y181C double mutant reverse transcriptase were computationally investigated. Quantitative structure-activity relationship (QSAR) studies, metabolic studies, and docking simulations were performed on these compounds. The logP value and the HOMO-LUMO energies of the compound were found to be critical for activity. The most probable biotransformation reactions include N5 / N11-dealkylation, aliphatic hydroxylation/oxidation at the ethoxy connector, and aromatic hydroxylation in the dipyridodiazepinone nucleus and the aryl group. Docking studies showed that the inhibitor must have at least two aromatic structures to maintain interactions with the Tyr181 – Tyr188 pocket and the Tyr318 region. The nucleus of the molecule must be a hydrophobic structure to interact with Val179 and Gly190 and must be substituted with H-bonding functional groups to interact with Lys101 and Lys/Asn 103.
INTRODUCTION
Acquired Immunodeficiency Syndrome (AIDS) is the disease caused by the human immunodeficiency virus (HIV). HIV contains its genome on RNA strands and for it to insert the genetic material in the genome of the host, a DNA copy of the single-stranded RNA should first be synthesized. This reversal of the transcription process is performed by the enzyme reverse transcriptase. Reverse transcriptase is a multifunctional enzyme with an RNA-directed DNA polymerase, DNA-directed DNA polymerase, and an RNAse H activity. The three dimensional conformation of the enzyme consist of a large claw-shaped active site. It is a heterodimer with a 66-kDa (p66) and a 51-kDa (p51) subunit, which are coded by the same gene (Kohlstaedt et al. 1992; Jacobo-Molina et al. 1993; Smerdon et al. 1994; Tantillo et al. 1994; Ding et al. 1995; Ren et al. 1995).
REFERENCES
CRUCIANI G, ARISTEI Y, VIANELLO R, BARONI M. 2005. GRID-Derived Molecular Interaction Fields for Predicting the Site of Metabolism in Human Cytochromes. In: Molecular Interaction Fields: Applications in Drug Discovery and ADME Prediction. Weinheim: WILE-VCH Ver lang GmbH and Co. KGaA. 310p.
DING J, DAS K, MOEREELS H, KOYMANS L, ANDRIES K, JANSSEN PA, HUGHES SH, ARNOLD E. 1995. Structure of HIV-1 RT/TIBO R 86183 reveals similarity in the binding of diverse nonnucleoside inhibitors. Nature Struct Biol 407-415.
GOODSELL DS, MORRIS GM, OLSON AJ. 1996. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit 9: 1-5.
GUEX N, PEITSCH MC. 1997. Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997. 18: 2714-23.
JACOBO-MOLINA A, DING J, NANNI RG, CLARK AD JR, LU X, TANTILLO C, WILLIAMS RL, KAMER G, FERRIS AL, CLARK P, ARNOLD E. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double stranded DNA at 3.0A° resolution shows bent DNA. Proc Natl Acad Sci 90: 6320-24.
JACOBS A, SEN J, RONG L, CAFFREY M. 2005. Alanine scanning mutants of the HIV gp41 loop. The J Biol Chem 280: 27284-88.
KIER LB, HALL LH. 1999. Molecular Structure Description: The Electrotopological State; Lemont B. Ed. New York: Academic Press. 286p.
KOHLSTAEDT LA, WANG J, FRIEDMAN JM, RICE PA, STEITZ TA. 1992. Crystal structure at 3.5A° resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256: 1783-90.
KOPP EB, MIGLIETLA JJ, SHRUTKOWSKI AG, SHIH CK, GROB PM, SKOOG MT. 1991. Steady state kinetics and inhibition of HIV-1 reverse transcriptase by a non-nucleoside dipyridodiazepinone, BI-RG-587, using a heteropolymeric template. Nucleic Acids Res 19: 3035-39.
MAI A, ARTICO M, RAGNO R, SBARDELLA G, MASSA S, MUSIU C, MURA M, MARTURANA F, CADEDDU A, MAGA G, LA COLLA P. 2005. 5-alkyl-2alkylamino-6-(2,6-difluorophenylalkyl)- 3,4-dihydropyrimidin-4(3H)-ones, a new series of potent, broad-spectrum non nucleoside reverse transcriptase inhibitors belonging to the DABO family. Bioorg Med Chem 13: 2065-67.
MEISLER G, TARR D. 1995. Inorganic Chemistry 2nd Ed. New Jersey: Prentice Hall, Inc. 480p.
O’ MEARA J, YOAKIM C, BONNEAU P, BOS M, CORDINGLEY M, DEZIEL R, DOYON L, DUAN J, GARNEAU M, GUSE I, LANDRY S, MALENFANT E, NAUD U, OGILVIE W, THAVONEKHAM B, SIMONEAU B. 2005. Novel 8-Substituted Dipyridodiazepinone Inhibitors with a Broadspectrum of Activity against HIV-1 Strains Resistant to Non-nucleoside Reverse Transcriptase Inhibitors. J Med Chem 48: 5580-88.
PETTERSEN EF, GODDARD TD, HUANG CC, COUCH GS, GREENBLATT DM, MEN EC, FERRIN TE. 2004. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J Comput Chem 25(13): 1605-12.
REN J, ESNOUF R, GARMAN E, SOMERS D, ROSS C, KIRBY I, KEELING J, DARBY G, JONES Y, STUART D et al. 1995. High Resolution Structures of HIV -1 RT from four RT-inhibitor complexes. Nature Struct Biol 2: 293.
RICHMAN DD. 1993. Resistance of clinical isolates of human immunodeficiency virus to antiretroviral agents. Antimicrob. Agents. Chemother 37: 1207-13.
ROMERO DL, BUSSA, M, TAN CK, REUSSER F, PALMER JR, POPPE SM, ARISTOFF PA, DOWNERY KM, SO AG, RESNICK L, TARPLEY WG. 1991. Nonnucleoside reverse transcriptase inhibitors that potently and specifically block human immunodeficiency virus type 1 replication. Proc Natl Acad Sci U.S.A. 88: 8806-10.
SMERDON SJ, JAGER REN J, ESNOUF R, GARMAN E, SOMERS D, ROSS C, KIRBY I, KEELING J, DARBY G, JONES Y, STUART D. 1994. High resolution structures of HIV-1 RT from four RTinhibitor complexes. Nature Struct Biol 2: 293.
SMITH MB, HOSE BM, HAWKINS A, LIPCHOCK J, FARNSWORTH DW, RIZZO RC, TIRADORIVES J, ARNOLD E, ZHANG W, HUGHES SH, JORGENSEN WL, MICHEJDA CJ, SMITH RH JR. 2003. Molecular modeling calculations of HIV1 reverse transcriptase nonnucleoside inhibitors: Correlation of binding energy with biological activity for novel 2-aryl-substituted benzimidazole analogues. J Med Chem 46: 1940-47.
SPENCE RA, KATI WM, ANDERSON KS, JOHNSON KS. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science 267: 988-993.
SPSS INC. 2001. SPSS for Windows Release 11.0.1 Chicago; Illinois: SPSS Inc.
TANTILLO C, DING J, JACOBO-MOLINA A, NANNI RG, BOYER PL, HUGHES SH, PAUWELS R, ANDRIES K, JANSENN PA, ARNOLD E. 1994. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase, Implications for mechanisms of drug inhibition and resistance. J Mol Biol 243: 369-387.
WALLACE AC, LASKOWSKI RA, THORNTON JM. 1995. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8: 127-134.
WANG J, KOHLSTAEDT LA, CHIRINO AJ, FRIEDMAN JM, RICE PA, STEITZ TA. 1994. Structure of the binding site for nonnucleoside inhibitors of reverse transcriptase of human immunodeficiency virus type 1. Proc Natl Acad Sci 91: 3911-15.
WOLD S. 1991. Validation of QSAR’s. Quantitative Structure – Activity Relationship 10: 191-193.
YARCHOAN R, MITSUYA H, THOMAS RV, PLUDA JM, HARTMAN NR, PERNO CF, MAREZYK KS, ALLAIN JP, JOHNNS DG, BRODER S. 1989. In vitro activity against HIV and favorable toxicity profile of 2’,3’-dideoxyinosine. Science 215: 412-415.
ZAMORA I, AFZELIUS L, CRUCIANI G. 2003. Predicting Drug Metabolism: A Site of Metabolism Tool Applied to the Cytochrome P450 CYP2C9. J Med Chem 46: 2313-24.









